New imaging method enables identification of rare synapses in the brain

The complexity of living systems is the result of intricate networks in which different biomolecular processes are precisely coordinated to form cells, tissues and entire organisms. To gain a comprehensive understanding of this biological complexity, the individual biomolecules must be identified, their occurrence in the body and their interactions with each other determined. Capturing this complexity with nanometer resolution for several molecules simultaneously is a current scientific and technological challenge. The vast majority of cellular functions are carried out by proteins. Proteins are biomolecules that are responsible for the shape and structure of cells as well as for biochemical metabolic processes. Until now, there was no technology that could easily recognize more than 15 protein species at a very high resolution.
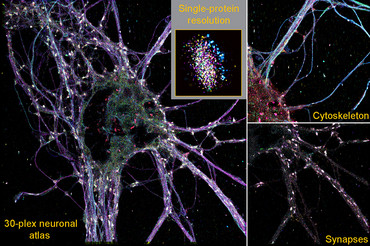
Scientists led by Dr. Eugenio F. Fornasiero, research group leader at the Department of Neuro- and Sensory Physiology at the University Medical Center Göttingen (UMG), Dr. Felipe Opazo, research group leader at the Department of Neuro- and Sensory Physiology and at the Center for Biostructural Imaging of Neurodegeneration (BIN) at UMG, Professor Dr. Ralf Jungmann, head of the "Molecular Imaging and Bionanotechnology" research group at the Max Planck Institute of Biochemistry (MPIB) in Martinsried and holder of the Chair of Molecular Physics of Life at the Ludwig-Maximilians-Universität München (LMU), and Dr. Carsten Marr, Director of the Institute of AI for Health at Helmholtz Munich, , have jointly developed a new high-resolution imaging technique that can be used to study the diversity of brain synapses. Synapses are the contact site between nerve cells and are used to transmit information. With their newly developed technology, the so-called SUM-PAINT method, the researchers were able to simultaneously display 30 different proteins in the nerve cells of the brain in a high-resolution three-dimensional structure (3D) and analyze their interaction. This form of simultaneous observation of molecules, also known as multiplexing, made it possible to decode the complexity of the synaptic protein composition of almost 900 synapses and identify a new type of rare synapse in the brain. The results have been published in the renowned scientific journal Cell.
Original publication:
Spatial Proteomics at single-protein resolution. Eduard M. Unterauer*, Sayedali Shetab Boushehri*, Kristina Jevdokimenko*, Luciano A. Masullo, Mahipal Ganji, Shama Sograte-Idrissi, Rafal Kowalewski, Sebastian Strauss, Susanne C.M. Reinhardt, Ana Periodic, Carsten Marr, Felipe Opazo, Eugenio F. Fornasiero§, Ralf Jungmann§. 2024 Cell 187, 1-16, March 28. https://doi.org/10.1016/j.cell.2024.02.045
* = shared first authors, § = correspondence
"The SUM-PAINT method can be carried out relatively easily with commercially available microscope hardware and provides a detailed overview of the localization and interaction of a large number of proteins at the molecular level. This makes it possible to investigate previously elusive details of neurological disorders and could contribute to a deeper understanding of the underlying disease mechanisms," says Dr. Eugenio F. Fornasiero from the UMG and one of the last authors of the study. "I am convinced that this technology will be instrumental in exploring previously unknown changes in the nervous system that lead to neurodegenerative diseases," says Dr. Fornasiero.
"This is a technological leap forward in super-resolution microscopy. We can capture the spatial arrangement and interaction of 30 proteins or other cell components simultaneously in one image. By combining several types of information in a single image, a more comprehensive picture of a sample can be obtained without having to take separate images for each feature. This enables a more efficient analysis of biological samples up to a hundred times faster than with previous microscopy techniques," says Eduard Unterauer, first author of the study from Prof. Jungmann's group.
Details of the Study
The study of protein composition has been greatly advanced by super-resolution microscopy techniques. However, many of these techniques cannot image more than a handful of target molecules at high resolution. The secondary label-based unlimited multiplexed (SUM)-PAINT method is based on the DNA-PAINT method developed by Prof. Dr. Ralf Jungmann's research group. This is a super-resolution technique that makes structures smaller than 20 nanometers visible. This is around 2,500 times smaller than the diameter of a human hair. Short dye-labeled DNA strands are used, which temporarily bind to their target molecules and produce a "blink", like a small sparkle in the dark. This blinking behaviour allows the target molecules to be visualized and localized in the cell. This information is combined to produce the super-resolution images.
In order to test the possibilities of SUM-PAINT and apply it to a cellular context, the groups of Dr. Fornasiero and Dr. Opazo at the UMG worked closely together to test, optimize and refine the strategies for reliable labelling of proteins in nerve cells. The use of antibodies and the much smaller nanobodies was of particular importance for the visualization of proteins in nerve cells. These were marked with short DNA codes and made it possible to specifically recognize and localize certain proteins in neurons. Analogous to DNA-PAINT, the position and structure of the target proteins are made visible by "blinking". This optimization work was the key to the first neuronal atlas, which characterizes 30 different proteins within neuorons. This was made possible thanks to the support of the Göttingen-based companies NanoTag Biotechnologies and Synaptic Systems.
In total, the complexity of the synaptic protein composition of almost 900 synapses was decoded. In order to examine the content-rich data sets, various analysis tools based on machine learning were developed and systematically used to analyze the data. In machine learning, artificial intelligence is trained to analyze data sets in order to derive hidden rules that would not be visible to the human eye. By extracting 1,600 features from the image data sets, such as protein content, distribution and shape, the researchers were able to discover a new, previously unknown type of synapse.
Kristina Jevdokimenko, a PhD student in the Fornasiero lab at UMG and also first author of the study, was able to show in brain tissue that this type of previously not seen synapses make up only about one percent of all synapses in the brain due to their unique protein composition and would not have been detected using other imaging techniques. The team has not only discovered a new type of synapse and provided unprecedented biological insight into synaptic diversity, but has also provided an integrated workflow for highly complex data generation and analysis. This is available to researchers around the world to gain an unprecedented understanding of their specific biological research questions.
CONTACT
University Medical Center Göttingen, University of Göttingen
Department of Neuro- und Sensory Physiology
Dr. Eugenio F. Fornasiero
Humboldtallee 23, 37073 Göttingen
Phone +49 551 / 39-67930
eugenio.fornasiero(at)med.uni-goettingen.de
fornasierolab.uni-goettingen.de
University Medical Center Göttingen, University of Göttingen
Center for Biostructural Imaging of Neurodegeneration (BIN)
Dr. Felipe Opazo
Von-Siebold-Str. 3a, 37075 Göttingen
Phone +49 551 / 39-61156
felipe.opazo(at)med.uni-goettingen.de
opazolab.de
University Medical Center Göttingen, University of Göttingen
Corporate Communication
Lena Bösch (Press contact UMG)
Von-Siebold-Str. 3, 37075 Göttingen
Phone +49 551 / 39-61020, Fax +49 551 / 39-61023
presse.medizin(at)med.uni-goettingen.de
www.umg.eu